Protein Engineering: Harnessing Machine Learning for Molecular Design
Protein engineering is a fascinating field at the intersection of biology, chemistry, and computer science. It involves the design and modification of proteins to create new or enhanced functionalities. In this project, I explored the potential of using advanced machine learning models to improve the process of developing new proteins with enhanced properties.
This project was my practicum for my master’s degree in analytics at Georgia Tech, where I applied my data science skills to the field of protein engineering. You can view the full technical report here.
The Challenge
The task was to develop a computational pipeline to efficiently identify superior protein variants for a diverse set of proteins. This challenge is crucial in many areas of biotechnology, from developing new enzymes for industrial processes to creating better therapeutics.
The main obstacles in protein engineering are:
- The high cost and time requirements of experimental testing. Ideally we can reduce the number of experiments needed to find the best protein variant. This also means we will typically have little or no functional data to start with.
- The complex relationship between protein sequence and function. This is where LLM type models can really shine. We need to be able to predict a protein’s function given its sequence.
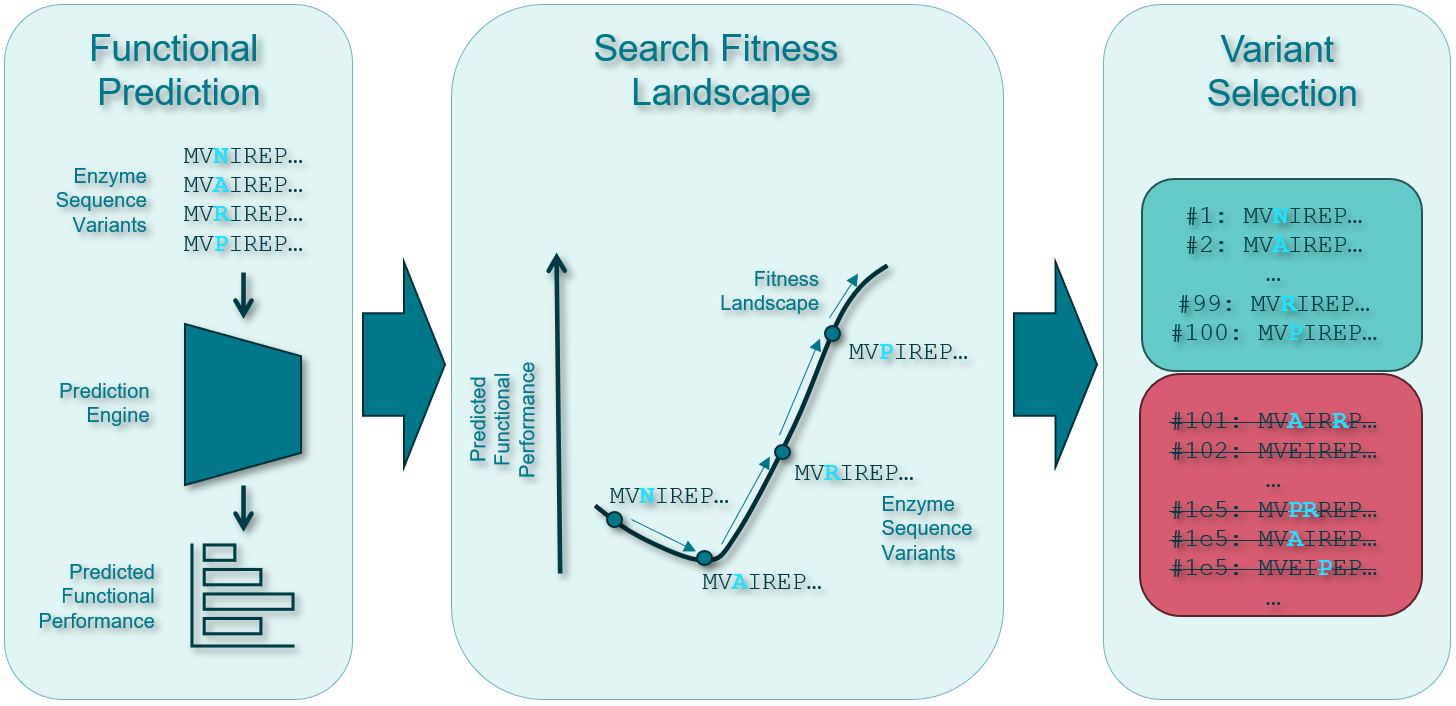
- The vast search space of possible protein sequences. This is like finding a needle in a haystack the size of the universe. This is called the “combinatorial explosion” problem. We need to find the optimal protein variants in this massive fitness landscape.
My goal was to use machine learning to navigate this complex landscape more efficiently, reducing the time and resources needed to discover improved protein variants.
The Method
For those interested in the technical details, here’s how I approached this challenge:
Data: I used a dataset of 34 deep mutational scanning (DMS) studies, spanning 25 organisms and various protein functions.
Zero-shot Learning: I employed the Evolutionary Scale Modeling (ESM) family of transformer-based protein language models developed by Facebook AI Research. These models can make predictions about protein function without any experimental data.
Few-shot Learning: Using the DMS experimental data, I trained various regression models (k-nearest neighbors, support vector machines, random forests, and ridge regression) on embeddings from the PLMs.
Gaussian Processes: To balance exploration and exploitation in my search for superior variants, I implemented Gaussian Process models that provide both predictions and uncertainty estimates.
Bayesian Optimization: I used a simplified Bayesian optimization approach to efficiently search the protein fitness landscape, leveraging the uncertainties provided by the GP models.
The Results
After developing and testing my computational pipeline, I found some key insights.
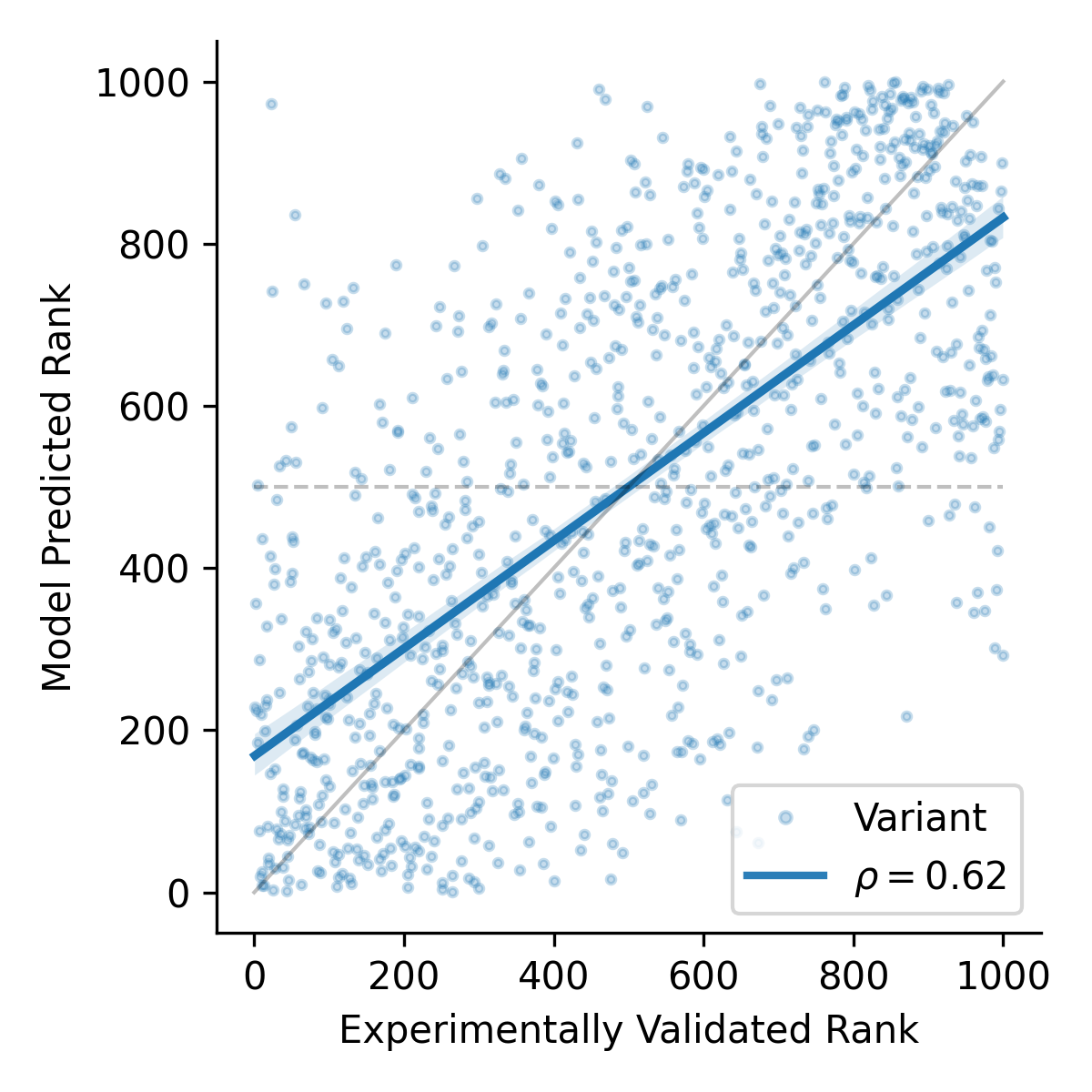
Zero-shot learning achieved a median Spearman rank correlation of ρ ≈ 0.52 between predicted and actual protein functionality.
Zero-shot learning with protein language models (PLMs) can reduce the effort required to find superior protein variants by nearly 50%.
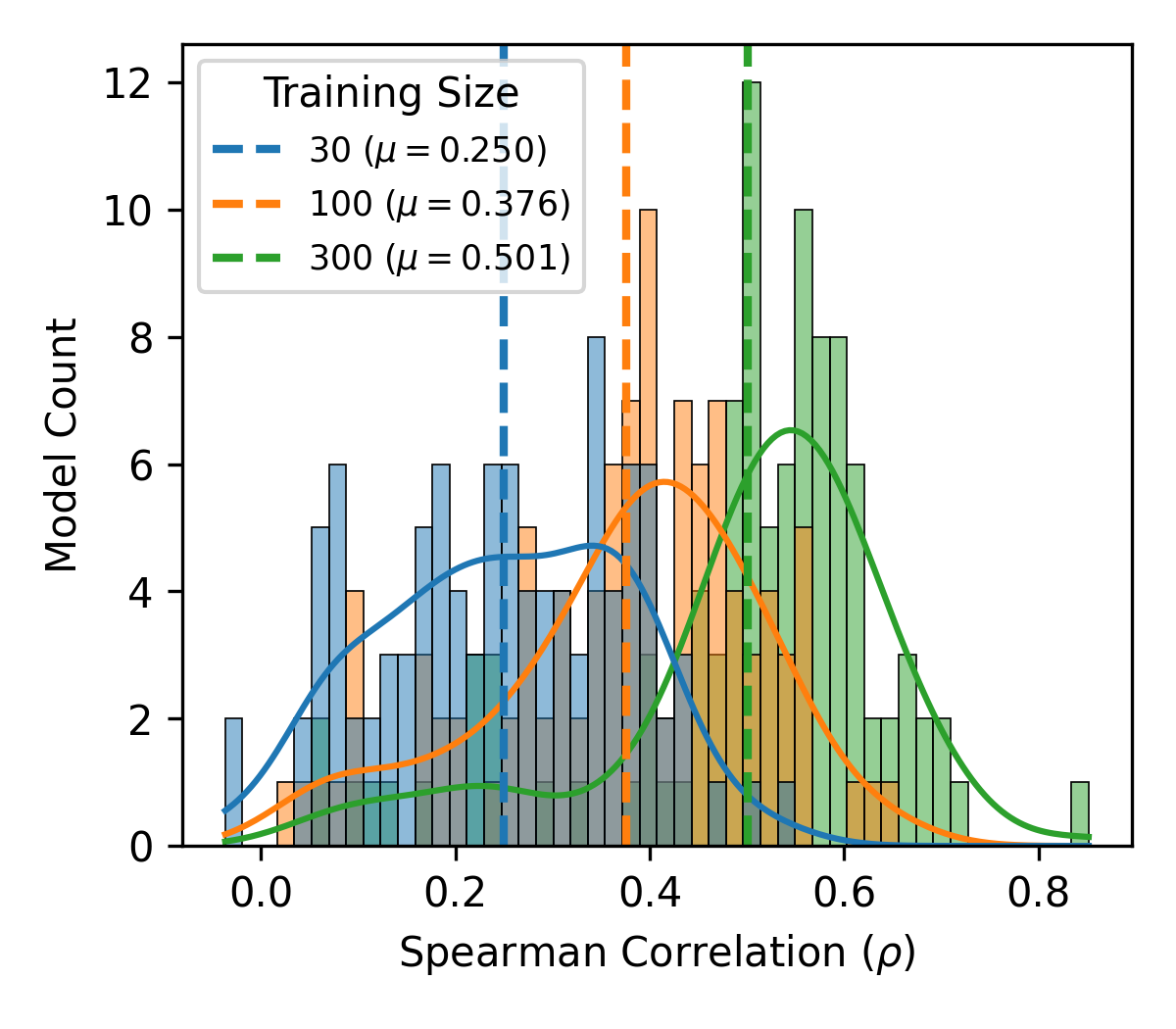
Few-shot learning with the simple regression algorithms under-performs zero-shot learning. This is likely due to the small number of data points available for training.
The Gaussian Process (GP) regression models start to outperform zero-shot predictions with more data, typically around 300 data points. The GP also outperforms the other few-shot simple regression models.
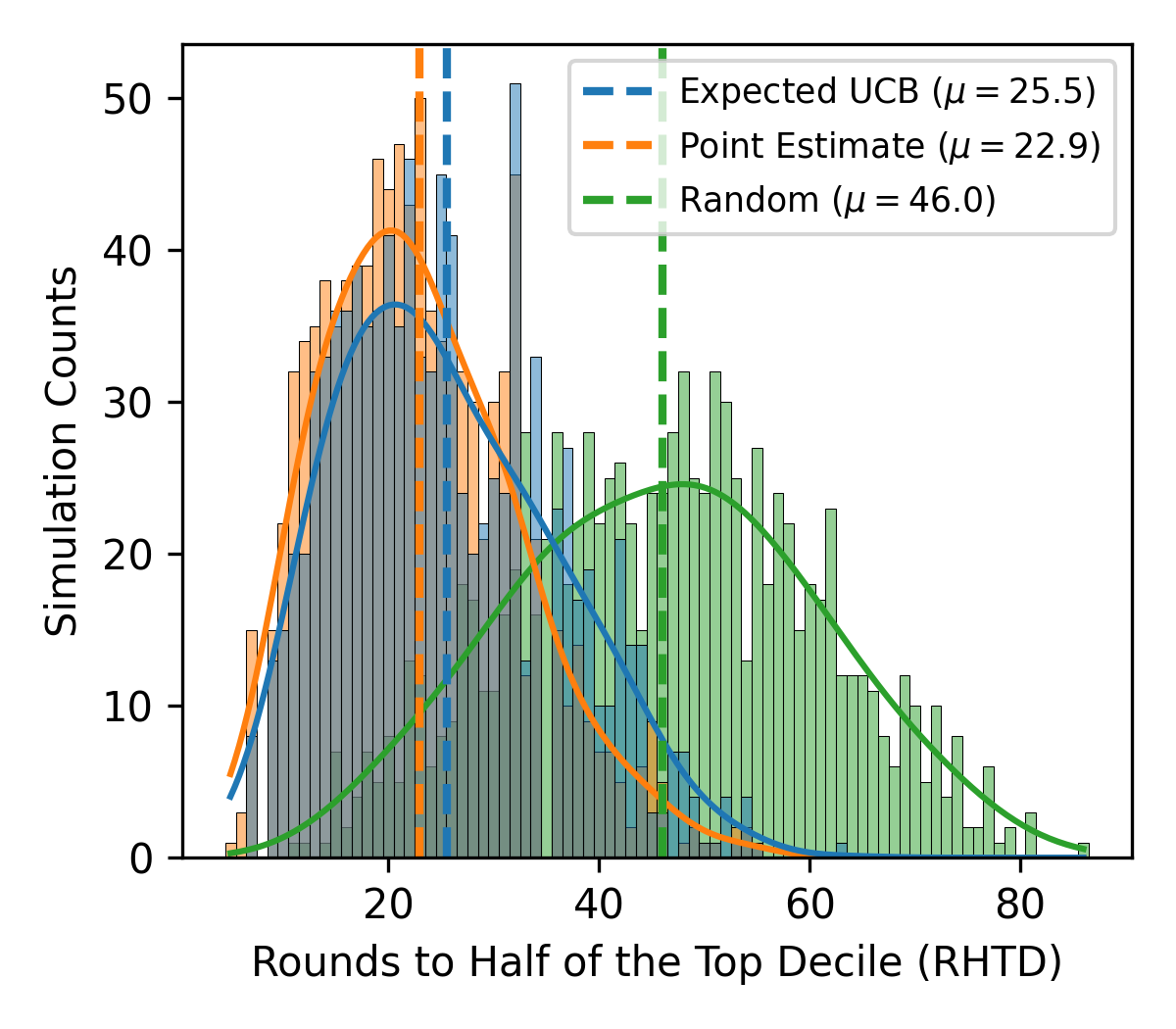
As modeled, using Bayesian Optimization (BO) with Gaussian Processes did not have a large improvement over point estimates. This bears further investigating.
What’s Next?
Having completed this project, there are several exciting directions to explore:
- Expand the approach to a wider range of proteins and functions, potentially uncovering general principles of protein engineering.
- Integrate structural information into the models to improve prediction accuracy.
- Develop more sophisticated Bayesian optimization strategies for navigating the fitness landscape.
- Apply this pipeline to real-world challenges in enzyme engineering or therapeutic protein design.
Recently the ProteinGym was launched. It is a website that contains a collection of benchmarks and leaderboards for comparing models that predict protein mutation effects. It is the protein equivalent of Papers With Code. It is also a source of more mutational data, with DMS substitutions and indels, as well as mutational clinical data.
Technical Details
For those who want to dive deeper into the nitty-gritty details, here is the Practicum Final Report. It includes all the mathematical formulas, model architectures, and performance metrics for a comprehensive understanding of the project.
This project demonstrates the power of machine learning in accelerating protein engineering. By combining state-of-the-art language models with clever optimization strategies, we can dramatically reduce the time and cost associated with developing new protein variants. As these techniques continue to evolve, they promise to revolutionize fields from industrial biotechnology to personalized medicine.